In this page, we provide a general overview of how the reactions catalyzed by DNAzymes take place. Bear in mind that the experimental designs vary from one laboratory to another, as well as the choice of substrates, and the way they are activated. Therefore, we recommend that you look up the primary reference for each specific DNAzyme.
RNA cleavage |
The first described RNA cleaving DNAzyme was able to cleave a DNA oligonucleotide at a position in the sequence where a single ribonucleotide was embedded. RNA cleaving DNAzymes are one of the most pursued kinds of DNA catalysts, in part due to their practical and analytical applications, nowadays being able to cleave an all-RNA substrate practically at any desired position. The cleavage reaction, that proceeds in the presence of divalent metal ions, forms a 5’ product bearing a 2’3’-cyclic phosphate terminus and a 3’ product bearing a 5’-OH terminus. Diverse ribonuclease protein enzymes, for instance, RNAse A, as well as ribozymes, such as the hammerhead, yield the same reaction products. 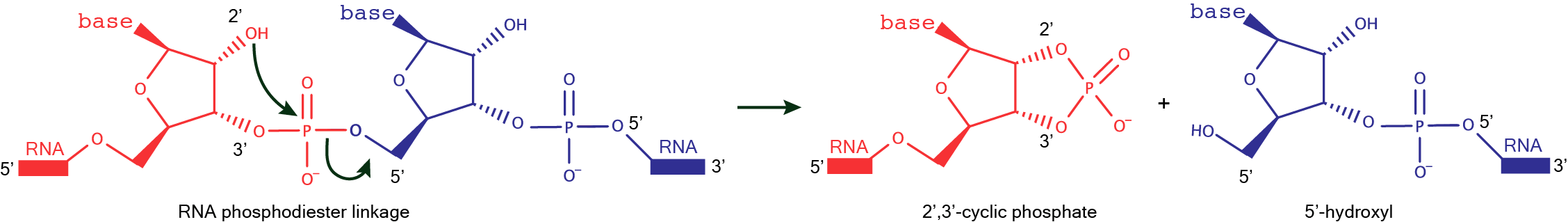 |
DNA cleavage |
DNAzymes can catalyze DNA cleavage either by DNA hydrolysis or by DNA deglycosylation and strand scission via two β-elimination reactions. Natural counterparts of these DNAzymes are DNases, that catalyze the hydrolytic cleavage of the DNA backbone; and DNA glycosylases, that remove damaged nucleobases from DNA and initiate the base excision repair pathway. 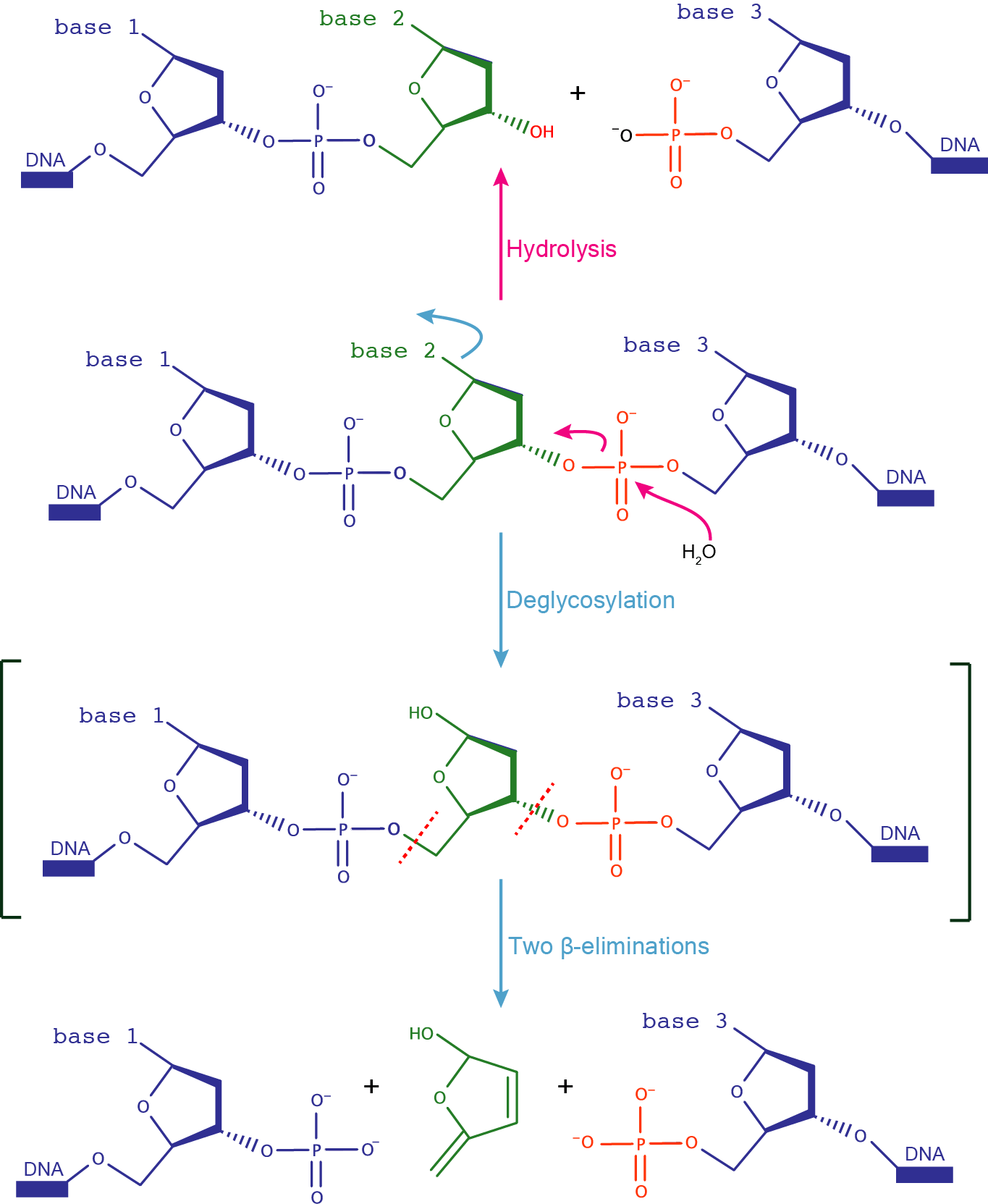 |
Covalent Modification of Amino Acid Side Chains |
The first successful DNAzyme-catalyzed reaction between a peptide side chain and an oligonucleotide exploited a three-helix junction organization that could bring together the peptide side chain and the electrophile (RNA 5'-triphosphate) for the reaction to take place. This strategy allowed for the identification of DNAzymes capable of forming a covalent bond between a substrate containing a single tyrosine residue (aromatic -OH group) and later a single serine residue (aliphatic -OH group) (A). Subsequent efforts focused on the identification of DNAzymes that could interact with a tripeptide substrate linked through a tether of varying length to a DNA anchor oligonucleotide (B), which incidentally revealed deoxyribozymes that could also catalyze reactions of free peptide substrates (C). For a more comprehensive overview on protein modification with DNAzymes, follow this link. 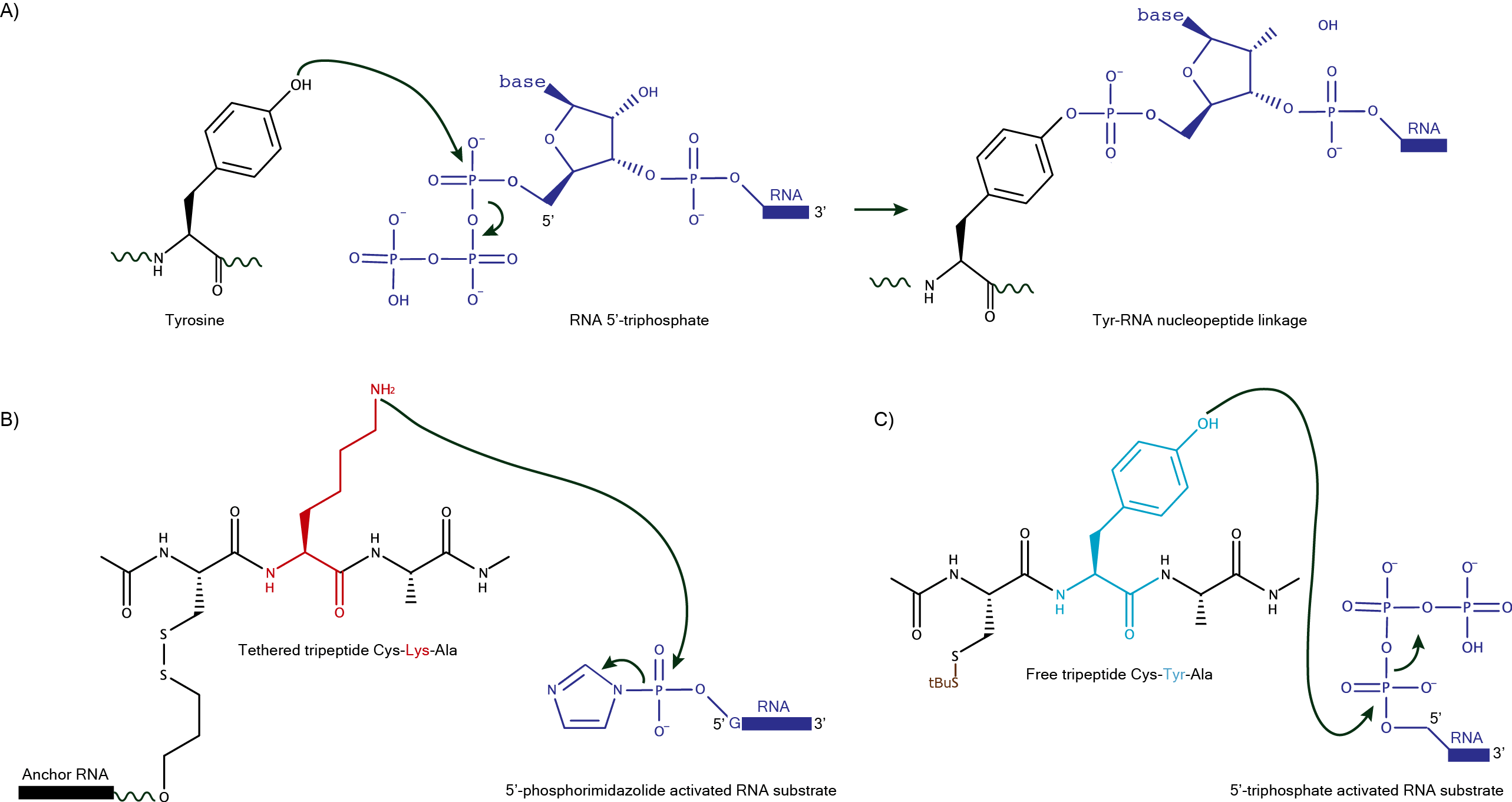 |
Thymine dimer repair |
Pyrimidine dimer formation in DNA results from exposure to UV light and represents a major kind of damage for the genetic material of living organisms, which can be carcinogenic or even lethal. This damage can be repaired by photolyase enzymes, which directly carry out photochemical repair (photoreactivation) the covalent dimers formed upon UV-light exposure. Prototypical DNAzymes carrying out this reaction include serotonin-dependent Sero1C and cofactor-independent UV1C. Intensive studies have led to the conclusion that the core of UV1C consists of G-quadruplexes positioned close to the thymine-dimer substrate, and it is believed that the electron transfer within the G-quadruplex would be responsible for the dimer's photoreactivation, similarly to the electron transfer occurring from flavin cofactors (in natural photolyase enzymes) to pyrimidine dimers within damaged DNA substrates. 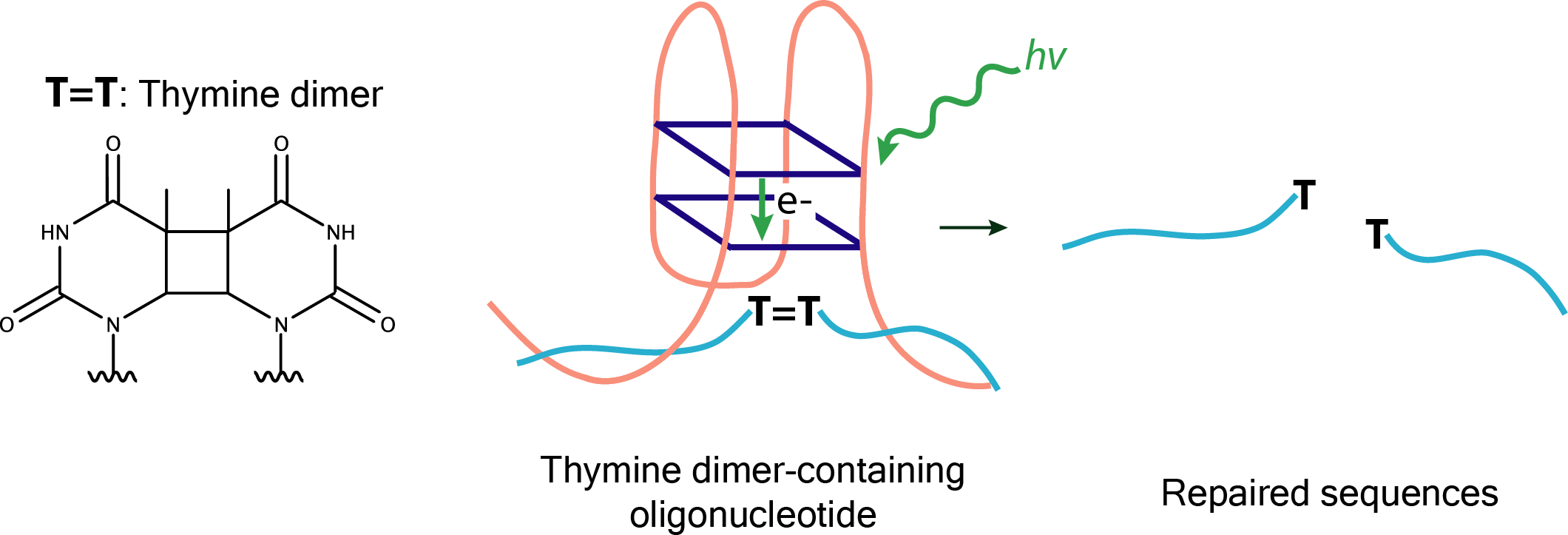 |
Porphyrin metalation |
Insertion of metal ions into porphyrins is called porphyrin metalation. Metalloporphyrins are essential in many cellular processes, for instance, in humans, ferrochelatase inserts Fe2+ into protoporphyrin IX, which is the final step in heme biosynthesis. DNAzymes are also capable of metalating porphyrin and can use a variety of ions as substrates. Many of the porphyrin-metalating DNAzymes are G-rich, which makes them particularly suitable for sensing heavy metals, such as Pb2+, due to their high affinity for G-quadruplex structures. |
DNA phosphorylation |
Analogous to polynucleotide kinases, which are widely used for the 5'-phosphorylation of DNA and RNA oligonucleotides, DNAzymes capable of self-phosphorylation have been isolated to make use of NTPs or dNTPs as a source of activated phosphate, and to act in the presence of different metal ion cofactors. In addition, although no natural enzymes are known to 3'-phosphorylate RNA or DNA termini, DNAzymes capable of 3'-phosphorylation of DNA substrates have been isolated. |
DNA capping |
DNA capping DNAzymes catalyze the transfer of the AMP of ATP to the phosphate group located at its 5' terminus, creating a 5’,5’-pyrophosphate cap. T4 DNA ligase catalyzed DNA ligation forms the same pyrophosphate cap structure. 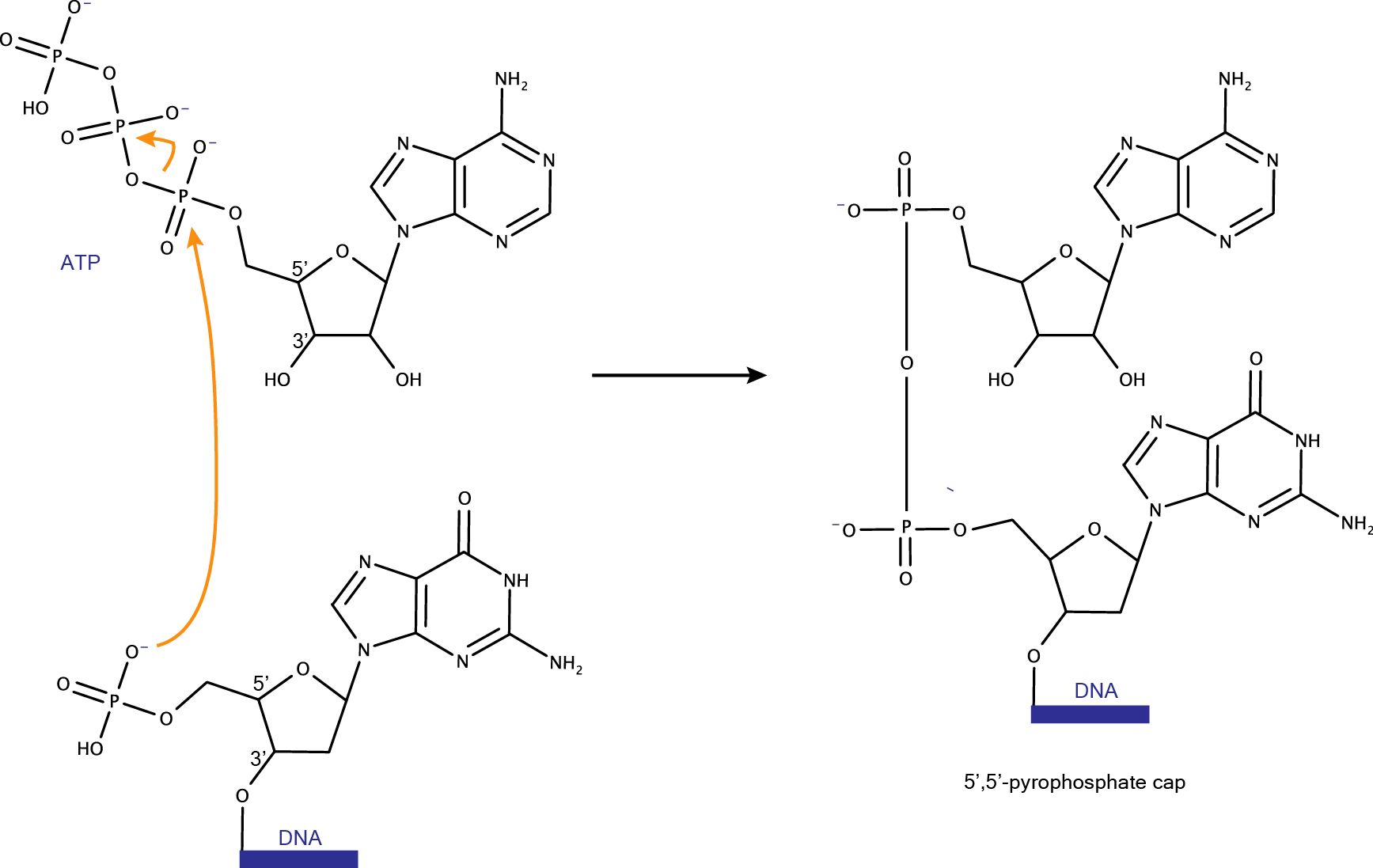 |
RNA ligation |
RNA-ligating DNAzymes can give different reaction products: linear RNA (native and non-native linkages), branched RNA (branched or lariat if the two substrates are connected), as well as some unnatural linkages. These DNAzymes are useful for the preparation of long RNAs for structural and functional studies. Different combinations of activated RNA substrates together with the appropriate DNAzyme can be used to achieve the desired ligated product. DNAzymes for the formation of RNA linear linkagesFor the formation of RNA native linkages, it is possible to join a 2’,3’-cyclic phosphate with a 5’-OH, or else, a 2’,3’-diol with a 5’-triphosphate. Using a 3’-OH group and a 5’-triphosphate, proteinaceous RNA polymerases, assisted by divalent cations, as well as the class I ligase and the L1 ligase ribozymes, catalyze the formation of a native RNA phosphodiester linkage. 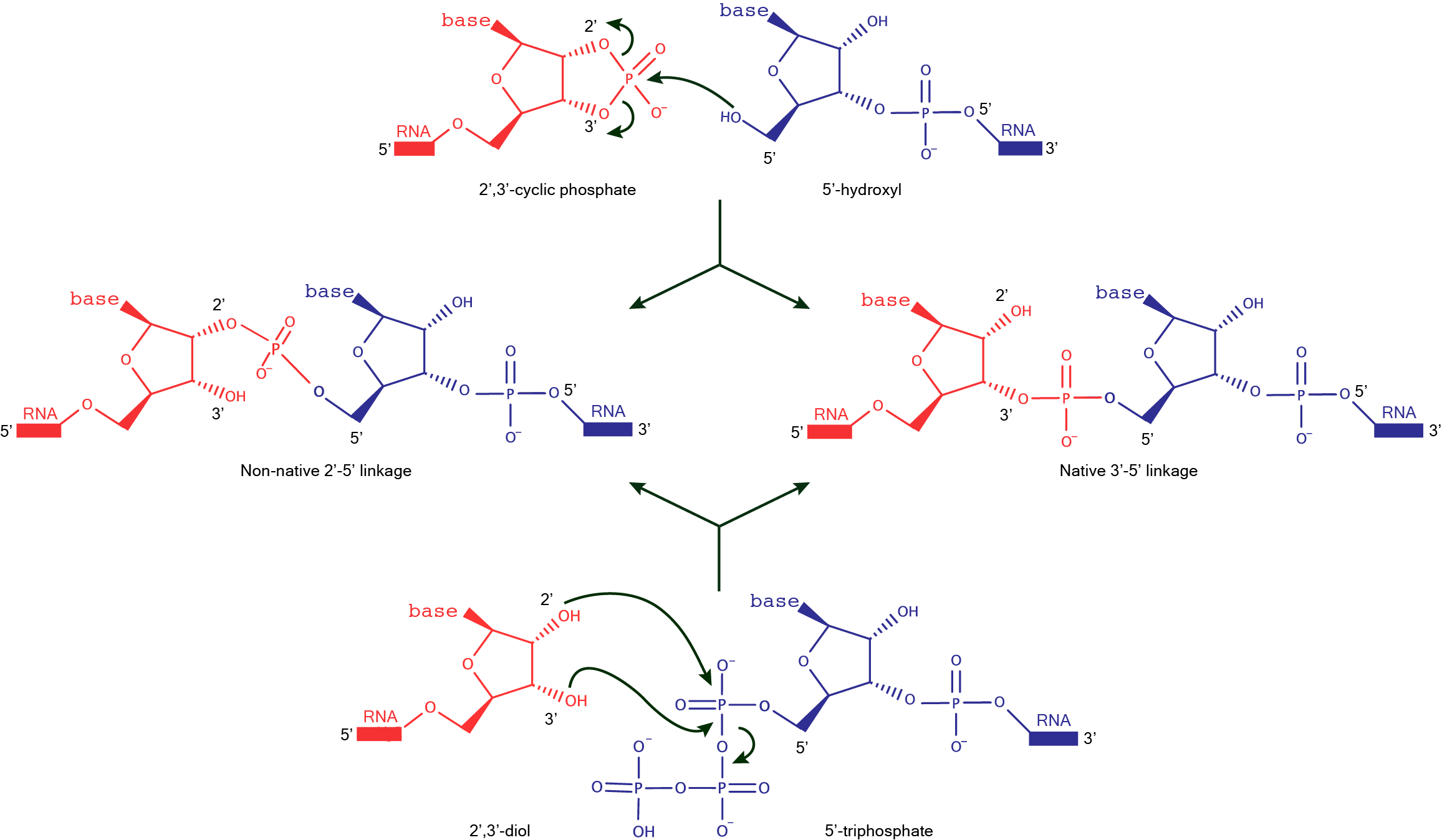 DNAzymes for the formation of branched and lariat productsThe preparation of branched or lariat RNAs, most commonly can be achieved when a 5'-triphosphorylated ribonucleotide attacks a 2’-OH group at the branching site. Natural 2',5'-branched RNA products are formed during pre-mRNA processing by the spliceosome. 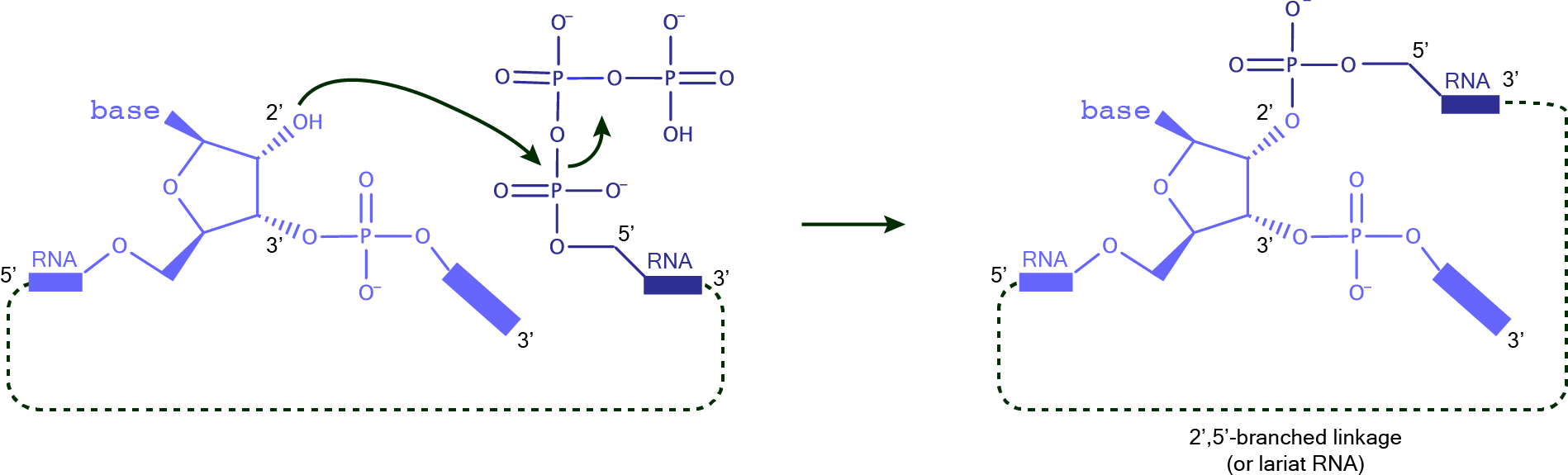 |
DNA ligation |
DNAzymes for the formation of DNA linear linkagesDNAzyme catalyzed formation of a native DNA linkage can be achieved upon condensation of the 5′–hydroxyl of one DNA substrate and the 3′-phosphorimidazolide of another DNA substrate. An alternative strategy requires, in the first place, the formation of a cap structure on the donor substrate (identical to that generated by T4 DNA ligase), and its subsequent reaction with the 3'-OH of the acceptor substrate to form the 3',5'-phosphodiester bond. 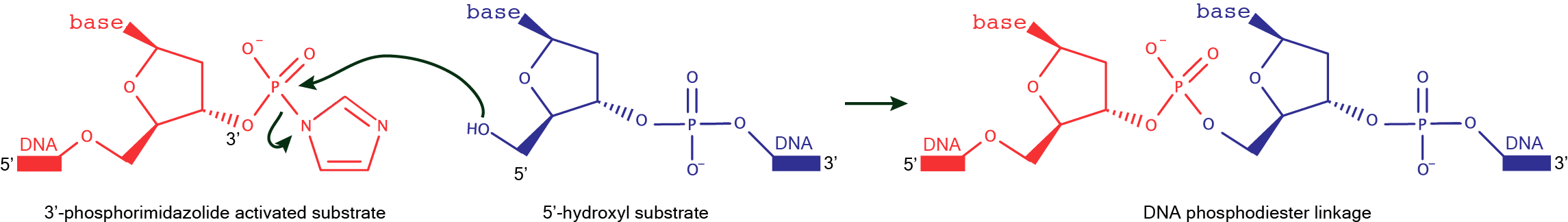 DNAzymes for the formation of branched DNADNAzyme catalyzed formation of a branched DNA is possible by joining the 2′-hydroxyl of the branch-site ribonucleotide of a DNA substrate to the activated 5′-phosphorus of a separate DNA strand. While the 5'-phosphorus is activated as a 5'-triphosphate for the synthesis of branched RNA, it is activated as a 5'-adenylate for the synthesis of branched DNA. These DNAzymes make use of a three-helix junction architecture, and examples of this family include 7S11 for the formation of branched RNA and 15HA9 for the formation of branched DNA. 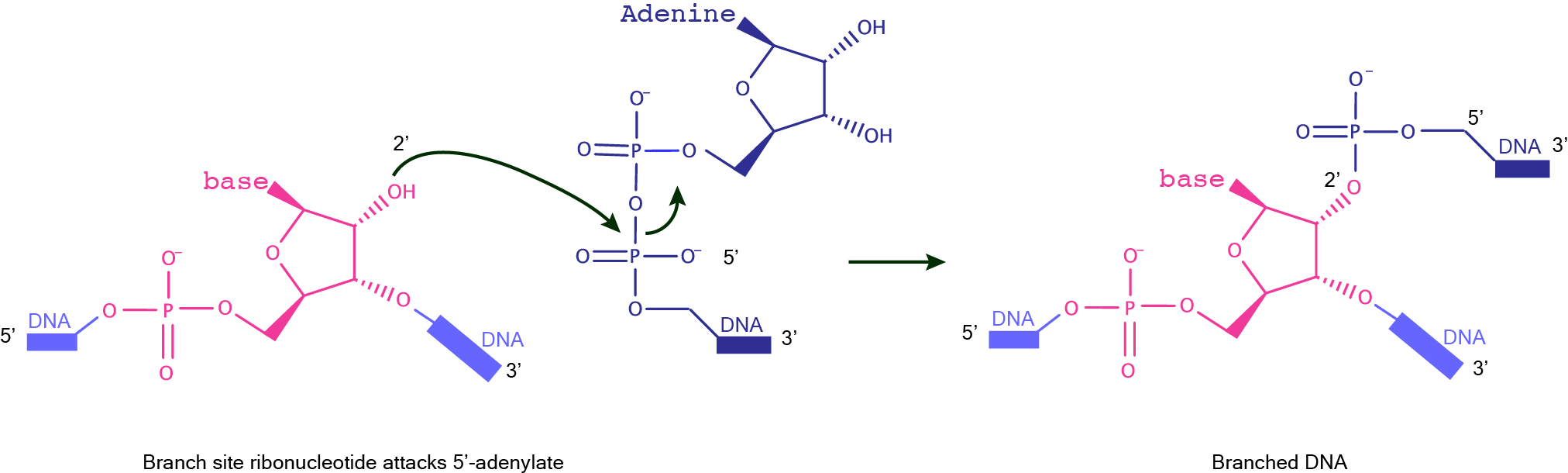 |
DNA depurination |
DNA depurination is a chemical reaction in which the β-N-glycosidic bond is hydrolytically cleaved releasing a purine nucleic base (adenine or guanine). The resulting abasic site is prone to strand scission, which is one of the mechanisms exploited by some DNA cleaving DNAzymes. The one example included to date in DNAmoreDB of a site-specific DNA depurinating DNAzyme is that of 10FN10. This DNAzyme self-depurinates position 1 in its sequence (see cheme below), and requires periodate as cofactor. Cofactor-independent site-specific deglycosylating DNAzymes are also known and will be shortly included in this database. 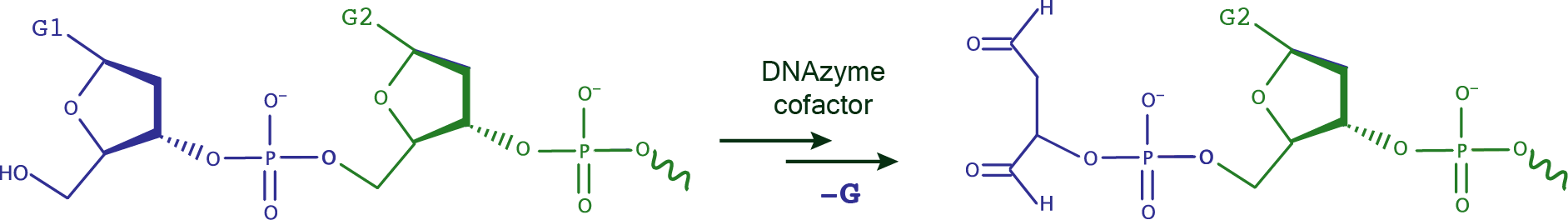 |